Phylogenetic tree POGIL answers key holds the key to unlocking the mysteries of evolutionary history. As we delve into this comprehensive guide, we’ll unravel the intricacies of phylogenetic trees, their construction, analysis, and practical applications, illuminating our understanding of the interconnectedness of life on Earth.
From the fundamental concepts to cutting-edge research, this exploration promises to empower you with a profound appreciation for the power of phylogenetic trees in deciphering the evolutionary narrative.
Phylogenetic Tree Basics
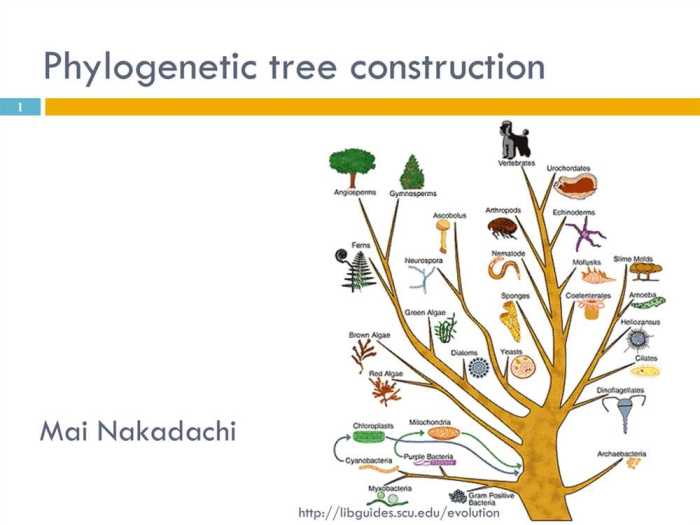
A phylogenetic tree is a diagram that depicts the evolutionary relationships between different species or other taxa. It is a branching diagram that shows the hypothesized evolutionary history of a group of organisms. The tree’s branches represent the different lineages that have evolved from a common ancestor, and the length of the branches represents the amount of evolutionary change that has occurred along each lineage.
Phylogenetic trees are important in evolutionary biology because they provide a visual representation of the relationships between different species. This information can be used to study the evolution of different traits, to infer the history of different groups of organisms, and to make predictions about the future evolution of a group.
Types of Phylogenetic Trees
There are two main types of phylogenetic trees: rooted and unrooted. Rooted trees have a single root node that represents the common ancestor of all the taxa in the tree. Unrooted trees do not have a root node, and the relationships between the taxa in the tree are not specified.
Rooted trees are more common than unrooted trees, and they are typically used to represent the evolutionary history of a group of organisms. Unrooted trees are sometimes used to represent the relationships between a group of organisms when the root node is not known.
Phylogenetic Tree Construction
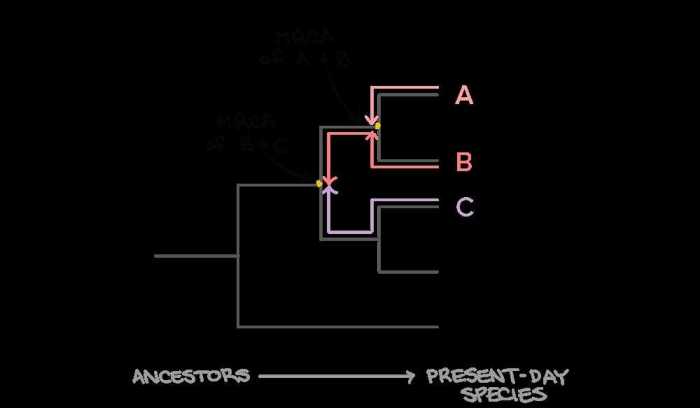
Phylogenetic trees are constructed using various methods, each with its advantages and limitations. Two commonly used methods are maximum parsimony and neighbor-joining.
Maximum parsimonyassumes that the simplest explanation for the observed data is the most likely. In this method, the tree with the fewest evolutionary changes, or mutations, is considered the most parsimonious and, therefore, the most likely to represent the true evolutionary history of the organisms.
Neighbor-joiningis a distance-based method that constructs a tree based on the pairwise distances between the organisms being studied. The algorithm starts by creating a star-shaped tree, with all organisms connected to a central node. It then iteratively joins pairs of organisms that are closest together, until a single tree is formed.
Example
Maximum parsimony has been used to construct a phylogenetic tree for the primates, based on DNA sequence data. The tree shows that humans are most closely related to chimpanzees and bonobos, and that these three species diverged from a common ancestor around 6 million years ago.
Neighbor-joining has been used to construct a phylogenetic tree for the bacteria, based on protein sequence data. The tree shows that the bacteria are divided into two main groups, the Gram-positive bacteria and the Gram-negative bacteria. The Gram-positive bacteria are further divided into two subgroups, the Firmicutes and the Actinobacteria.
Phylogenetic Tree Analysis: Phylogenetic Tree Pogil Answers Key
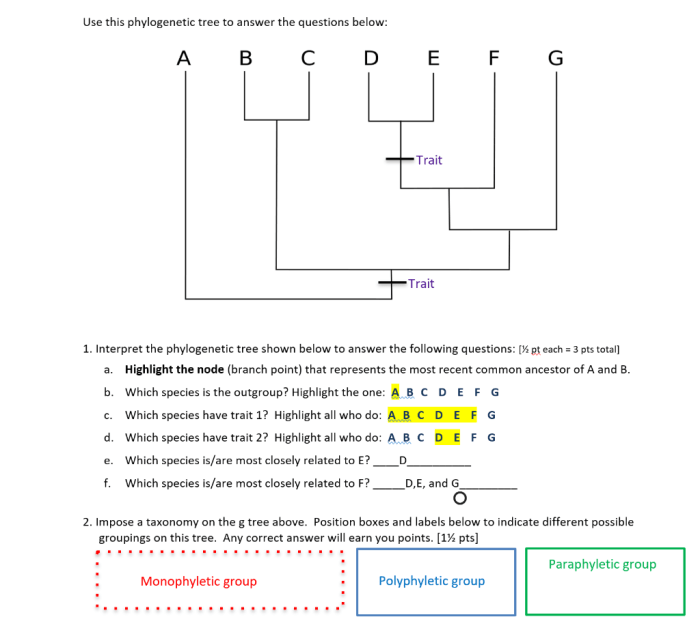
Phylogenetic trees provide valuable insights into the evolutionary relationships between species. They are constructed based on shared characteristics, which can be morphological, genetic, or a combination of both. By analyzing these trees, scientists can infer the evolutionary history of species and their common ancestors.Phylogenetic
trees have numerous applications in comparative genomics, where they are used to study the evolution of gene families, identify orthologs and paralogs, and infer gene function. Additionally, they are employed in fields such as ecology, conservation biology, and medicine to understand the distribution, adaptation, and disease susceptibility of species.
Comparative Genomics
Phylogenetic trees play a crucial role in comparative genomics by enabling researchers to:
- Identify orthologs: Genes that share a common ancestor and have similar functions across different species.
- Identify paralogs: Genes that arise from duplication events within a genome and may have diverged in function.
- Study the evolution of gene families: Trace the evolutionary history of gene families, including gene duplication, loss, and divergence.
- Infer gene function: Predict the function of genes in one species based on the known function of orthologs in other species.
Other Research Fields, Phylogenetic tree pogil answers key
Phylogenetic trees are also widely used in other research fields, including:
- Ecology:To study the evolution of ecological traits, such as habitat preferences and niche partitioning.
- Conservation biology:To identify endangered species and develop conservation strategies based on evolutionary relationships.
- Medicine:To understand the genetic basis of diseases and develop targeted therapies based on evolutionary insights.
Phylogenetic Tree Interpretation
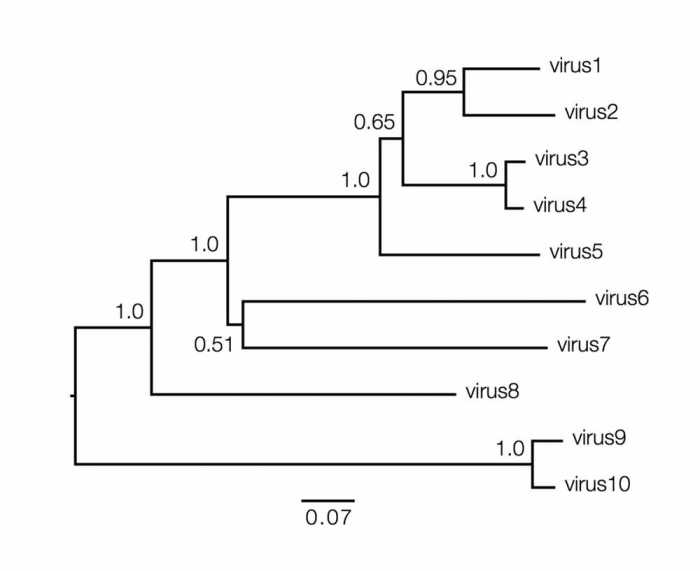
Phylogenetic trees are powerful tools for visualizing and interpreting evolutionary relationships among species. They provide a visual representation of the hypothesized branching order of species from a common ancestor. By studying phylogenetic trees, scientists can gain insights into the evolutionary history of different species, including their relatedness, divergence times, and patterns of evolution.Key
features of a phylogenetic tree include:
- Nodes:Nodes represent common ancestors of the species on the tree.
- Branches:Branches connect nodes and represent the evolutionary relationships between species.
- Root:The root of the tree represents the most recent common ancestor of all the species on the tree.
- Tips:The tips of the tree represent the extant species or taxa included in the analysis.
Phylogenetic trees can be interpreted to infer evolutionary relationships among species. For example, the length of a branch can be used to estimate the amount of evolutionary change that has occurred along that branch. The branching order of species can be used to infer the order in which species diverged from a common ancestor.Phylogenetic
trees have been used to make numerous scientific discoveries. For example, phylogenetic trees have been used to:
- Identify the closest living relatives of extinct species
- Trace the evolution of new species
- Understand the origins of diseases
- Develop new drugs and vaccines
- Conserve endangered species
Phylogenetic Tree Applications
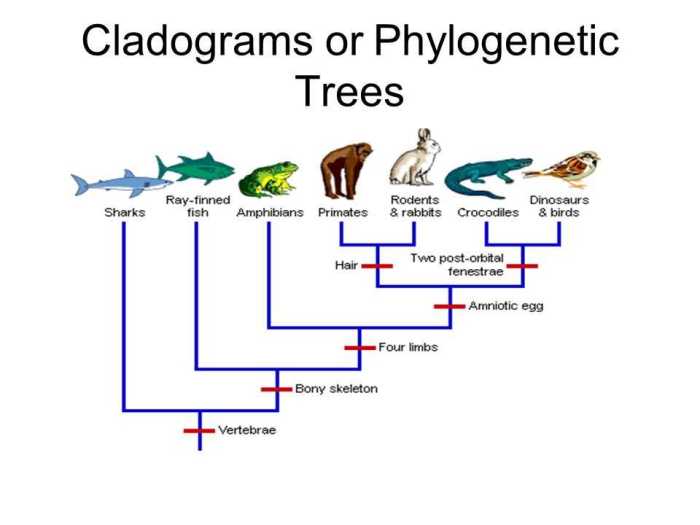
Phylogenetic trees are not only limited to the realm of evolutionary biology but also have significant practical applications in various fields. Their ability to depict evolutionary relationships has proven invaluable in disciplines such as medicine, agriculture, and conservation biology.
Medicine
In the medical field, phylogenetic trees have been instrumental in understanding the evolution and spread of infectious diseases. By constructing phylogenetic trees of pathogens, researchers can identify the origins of outbreaks, trace transmission routes, and predict the likelihood of future pandemics.
This knowledge has aided in the development of targeted vaccines and therapeutic interventions, ultimately improving human health and well-being.
Agriculture
Phylogenetic trees have revolutionized the field of agriculture by providing insights into crop domestication and improvement. By studying the evolutionary relationships among different plant varieties, scientists can identify desirable traits, such as disease resistance or drought tolerance, and incorporate them into new cultivars.
This approach has led to the development of hardier and more productive crops, contributing to global food security.
Conservation Biology
In the realm of conservation biology, phylogenetic trees play a crucial role in assessing the genetic diversity of endangered species. By understanding the evolutionary history and genetic relatedness among populations, conservationists can prioritize conservation efforts and develop strategies to maintain genetic diversity.
Phylogenetic trees also help identify genetically distinct populations that require specific conservation measures, ensuring the preservation of unique genetic lineages and the long-term survival of species.
Popular Questions
What is a phylogenetic tree?
A phylogenetic tree is a diagrammatic representation of the evolutionary relationships among different species or other taxa, based on shared characteristics and genetic similarities.
How are phylogenetic trees constructed?
Phylogenetic trees are constructed using various methods, such as maximum parsimony, neighbor-joining, and Bayesian inference, which analyze genetic data to infer evolutionary relationships.
What are the different types of phylogenetic trees?
There are different types of phylogenetic trees, including rooted trees (with a designated common ancestor) and unrooted trees (without a designated common ancestor), as well as cladograms (showing only branching patterns) and phylogram (showing both branching patterns and branch lengths).
What are the applications of phylogenetic trees?
Phylogenetic trees have wide-ranging applications in fields such as medicine, agriculture, and conservation biology, including studying disease transmission, identifying crop relatives for breeding, and assessing conservation priorities.